Genome engineering with HDR
Homology-directed repair (HDR) is a process where a DNA double-strand break (DSB) is repaired by homologous recombination using a DNA template. This template can come from within the cell during late S phase or G2 phase of the cell cycle, when sister chromatids are available prior to the completion of mitosis. Additionally, exogenous repair templates can be delivered into a cell, most often in the form of a synthetic, single-strand DNA donor oligo or donor plasmid, to generate a precise change in the genome.
Use of plasmid donor for HDR
Plasmid donor repair template is delivered into a cell to insert or change large sequences (knock-in a fluorescent reporter or replace an entire gene) of DNA in the endogenous genomic target region.
Design considerations for HDR using plasmid donors
To design a DNA donor plasmid for precise gene editing using HDR, one must consider:
- The distance of the intended HDR insertion site (or alteration site) from the Cas9 cut site
- The gRNA efficiency near a specific insertion site
- The length of homology arms
- Disruption of the CRISPR targeting site if it is present in the plasmid donor repair template
Here we describe designing HDR plasmid donors for use with the CRISPR-Cas9 system; general principles can be applied to any site-directed nuclease.
Insertion site and cut site distance
In mammalian cells, HDR has highest efficiency when the insertion site and the DSB are within ten nucleotides proximity. HDR efficiencies drop quickly as this distance increases1.
Selecting gRNAs for maximal HDR efficiency at a specific insertion site
Since HDR efficiency is relatively low compared to non-homologous end joining (NHEJ) repair of DSBs, it is important to choose a gRNA with the highest possible DNA cutting efficiency. An NHEJ-mediated efficiency of at least 25% is recommended. This is because the precise HDR-mediated repair will only occur in a fraction of DSBs that are available.
Additionally, not every insertion site will have PAM sequences within the surrounding 20 nt, or the assessed gRNA efficiency with available PAM sites might be very low. In these circumstances, choose the most active gRNA that is as close as possible to the intended insertion site. Again, be aware that HDR has highest efficiency when the insertion site and the DSB are within ten nucleotides from one another1.
Length of homology arms for a plasmid donor
Homology arm length for a plasmid donor typically ranges from 500 to 1000 nt2-4.
Design a plasmid donor
The following are basic design considerations that are automatically incorporated when using the HDR Donor Designer.
Retrieving a genomic sequence
An annotated genomic sequence is required to begin designing a plasmid donor. When selecting the genomic sequence, it is essential to retrieve the genomic sequence (introns and exons) and not, for example the mRNA sequence, so that your DNA template correctly repairs the intended HDR modification site.
Determine the insertion location
The insertion location should be within the endogenous gene of the target protein. Ideally, the insertion site should be precisely after the start codon for an N-terminal insertion, or conversely, immediately before the stop codon for a C-terminal insertion.
Primer design to amplify homology arms
When designing PCR primers to amplify the homology arms of your intended gene target, the chosen insertion site indicates the start location of the insert adjacent primers (Figure 1). The distance that the corresponding primer pair is away from the insertion site determines the length of the respective homology arm. Additionally, for proper ligation-free assembly into a plasmid donor, adapter sequences are appended to the gene specific portion of the primers (Figure 1, red or orange rectangles).
Homology arm primer design for generating a mKate2 donor plasmid in-frame with your target gene
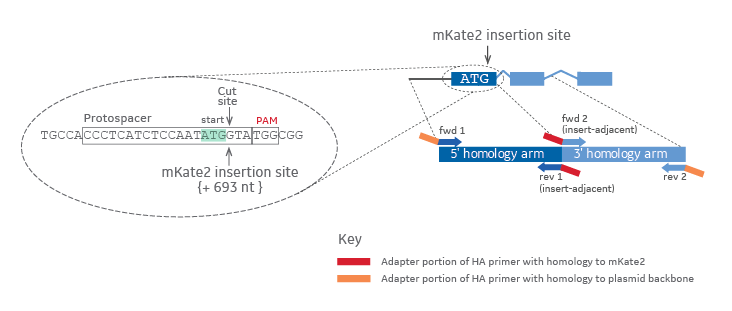
Figure 1. The gRNA target or Protospacer and PAM sequences are outlined in black in a zoomed in view on the left. The cut site and insertion site are indicated by arrows. The start codon (ATG) is highlighted in light green. Colors on the zoomed out diagram to the right indicate the origin of the DNA (dark blue = 5' homology arm, light blue = 3' homology arm, red = mKate2 fluorescent reporter sequence, orange = plasmid backbone).
Avoiding CRISPR-Cas9 cutting of a gene locus after HDR
When designing a donor plasmid, it is essential to disrupt the CRISPR target site when it occurs within your donor template. Failure to disrupt the CRISPR target site will allow Cas9 to cut your HDR-edited gene locus after the repair has taken place. This can be accomplished with one or more of the following changes to the donor template (Figure 2): (1) design the donor plasmid so that the DNA insertion splits the 20 nucleotide CRISPR target sequence, or a portion of the target sequence, and the corresponding PAM, or (2) introduce silent mutations in the PAM or CRISPR target region by introducing one or more nucleotide changes in the appropriate gene-specific insert-adjacent homology arm primer.
Design strategies to aviod CRISPR-Cas9 cutting after HDR-mediated insertion using a donor plasmid
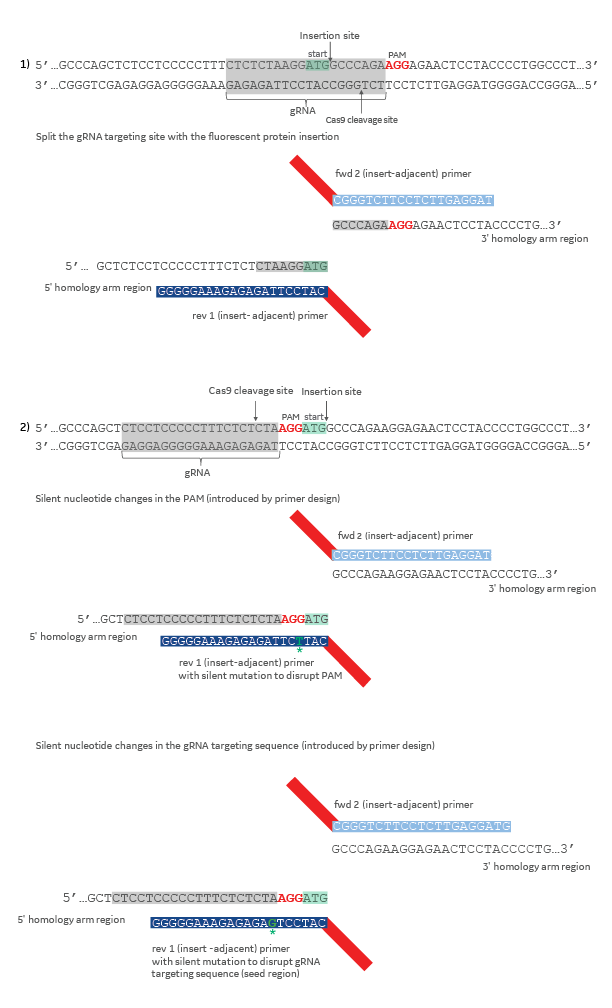
Figure 2. The gRNA target sequence is highlighted in gray and the PAM sequence in red. The start codon (ATG) is highlighted in light green. Silent mutations in primer sequences are colored dark green and indicated with an asterisk. Colors on the diagram indicate the origin of the DNA (dark blue = 5' homology, light blue = 3' homology, red = mKate2 fluorescent reporter sequence.
References
- B. Elliott, C. Richardson, et al. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 18(1), 93-101 (1998)
- R. Mahen, B. Koch, et al. Comparative assessment of fluorescent transgene methods for quantitative imaging in human cells. Mol. Biol. Cell. 25(22), 3610–3618 (2014)
- F. Merkle, W. Neuhausser, et al. Efficient CRISPR-Cas9-Mediated Generation of Knockin Human Pluripotent Stem Cells Lacking Undesired Mutations at the Targeted Locus. Cell Reports. 11(6), 875-883 (2015)
- M. Ratz, I. Testa, et al. CRISPR /Cas9-mediated endogenous protein tagging for RESOLFT super-resolution microscopy of living human cells. Scientific Reports. 5(9592),1-6 (2015)
Order Products
HDR Donor Designer
Quickly and easily design and order a donor template for precise CRISPR-Cas9 gene editing.
Edit-R HDR Plasmid Donor Kits
Rapidly and easily assemble a plasmid donor for HDR
Edit-R HDR Plasmid Donor Components
Quickly and efficiently build a HDR donor plasmid
Edit-R HDR Plasmid Donor Primers
PCR components for Edit-R Plasmid Donor Kits
CRISPR Design Tool
Design custom crRNA and sgRNA for gene editing applications
CRISPR-Cas9 Gene Editing
Optimized tools for high-confidence genome engineering
Helpful Resources
Read app note
Fluorescent tagging of an endogenous gene by homology-directed repair using Dharmacon Edit-R CRISPR-Cas9 reagents - Application Note Performing endogenous gene tagging using Dharmacon Edit-R Cas9 mRNA, synthetic crRNA-tracrRNA, and an EGFP donor plasmid.
Read app note
A suggested protocol optimizing delivery of the HDR reagents in difficult to transfect K-652 cells using an electroporation method.
