Using functional genomic screening to unveil mechanism of action: considerations of screen design
Reverse genetic screening allows researchers to understand gene function through observation of phenotype following gene modification. Thanks to the advent of techniques which allow genome-wide modification of gene expression, such as RNAi and CRISPR, massively paralleled screening of genes has unveiled the function of genes and their genetic networks, as well as to enable the identification of genetic elements involved in disease. This is broadly known as Functional Genomic Screening (FGS). This genetic methodology has allowed the elucidation of the mechanism of action (MoA) of exogenous agents which modulate intracellular gene function, such as pharmaceutical agents and other compounds.
The process of using FGS to unravel the MoA of a drug or compound is well established, and Revvity have extensive experience in providing the tools and services to facilitate these types of investigations (Figure 1).
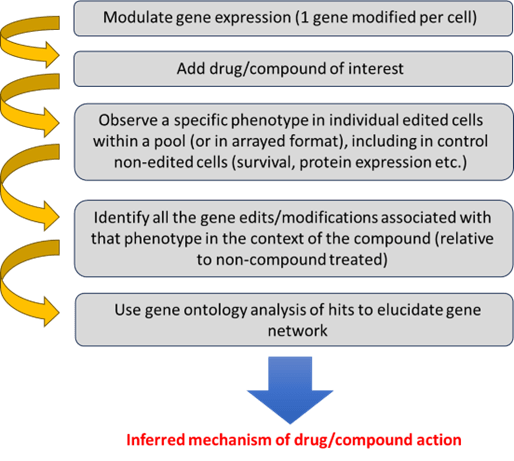
Figure 1. Scientific workflow to discern mechanism of action.
Five key considerations for CRISPR screening
1) There are many aspects of a screen that can be tailored to discern MoA. When a scientist has a novel compound they wish to investigate, the first consideration is the model system to select for experimentation. This model could be a cell line grown in 2D - such as an epithelial cancer cell derived from the diseased tissue of interest in which the compound is thought to target, or it could be a more complex 3D model – such as organoids. Organoids are groups of different cell types arranged in structures representing ‘mini-organs’ which can be grown in a three-dimensional matrix and are more representative of in vivo biological systems.
2) The second key consideration is which FGS technology to utilize. Loss of function technologies include CRISPRko (gene knockout), CRISPRi (gene expression interference) and siRNA (RNA silencing), and these allow the assessment of cellular phenotype following gene ablation or gene expression impairment. CRISPRko provides the knock-out of a gene, whereas for CRISPRi or siRNA, gene expression is knocked-down. This allows CRISPRi and siRNA to model druggability more closely and provides the researcher an orthologous approach to CRISPRko. Indeed, dual loss of function screening, where CRISPRko and CRISPRi screening is performed in parallel, can build confidence in understanding mechanism of action by confirmation of shared hits, as well as providing the advantage of identifying hits that are unique to each technology approach.
Alternatively, a gain of function approach can be achieved by CRISPRa (gene activation), and this allows a phenotype to be assessed in the presence of gene overexpression. Dual direction screening is when CRISPRa is paired with a loss of function technology (CRISPRko or CRISPRi). By combining parallel studies of gene ablation or gene expression impairment with overexpression, we can examine for opposite phenotypic effects on genes, as well as add extra dimensionality to pathway identification and insights into MoA. For example, Revvity have previously utilised all three CRISPR technologies (CRISPRko, CRISPRi and CRISPRa) for a genome wide screen to identify genes that are involved in both sensitivity and resistance to the BRAF inhibitor, vemurafenib (Dual CRISPRi/a poster). This approach allowed MoA elucidation through systematic hit identification and evaluation of gene networks in respect to the drug function.
If more complex network analysis is required, Revvity also offer dual-guide perturbation screens, where two genes of interest are perturbed in each cell simultaneously.
3) Thirdly, the scientist needs to consider if they wish to conduct a whole genome analysis using an off-the-shelf library, such as those offered by Revvity, or whether a more focused genetic screen is required with a targeted pool of selected candidates. The number of different guides targeting each individual gene also needs to be considered as this will have an effect on statistical power and confidence during hit calling and data analysis. For instance, statistical analysis of pooled screens which look for gene modifications that cause sensitivity to a drug, where those affected cells become depleted from the pooled population, can benefit from having a higher number of guides.
4) The fourth consideration for the scientist is to decide how compound function should be measured and what the functional readout is. This functional readout could be a survival/death phenotype, or something more nuanced such as the expression of particular protein markers in response to drug exposure.
5) The final consideration to help unveil MoA, is to decide whether the screening analysis will be conducted in an arrayed format – where the cells separated in each well of a multiwell plate are each treated with an individual gene modulator (guide RNA or siRNA), or whether a pooled screening format should be used – where all the candidates genes to be perturbed are modulated in a pool of cells, with each cell only receiving a single modulator. Arrayed screens can offer advantages of immediate phenotypic information, including the possibility of multiple phenotypic read-outs. They are also suitable for non-dividing cells such as neurons and co-culture systems where the scientist might want to measure the phenotype of a cell that is not being edited.
Pooled screens are generally suited to more simple readouts (for example, viability), they can provide an unbiased approach to whole genome screening and they use next-generation sequencing (NGS) as a read-out. Additionally, the complexity of a pooled screen readout can be increased through the use of technologies such as high content imaging (HCI) and fluorescence-activated cell sorting (FACS), where phenotypes can be investigated using antibody staining, or fluorescently tagged proteins.
Interpretation of the CRISPR screen
Once the screen has been conducted, the scientist then needs to correlate drug function with genetic perturbation and phenotypic readout. This can be achieved through candidate or ‘hit’ identification, gene network analysis, hit validation, and then hypothesis testing experimentation. See figure 2 for an example of how we might use FGS to investigate the MoA of a hypothetical novel therapeutic compound which induces cell death.
Summary
CRISPR screening is a powerful tool for helping to dissect the mechanism of action of a drug or compound. A variety of careful considerations should be taken when approaching these experiments depending on the phenotypic readout and the scope and scale of the investigation. Furthermore, by using a combination of different CRISPR technologies, such as those which can induce gene repression and overexpression in parallel experiments, more confidence can be taken when hit calling and when identifying genetic networks involved in drug function.
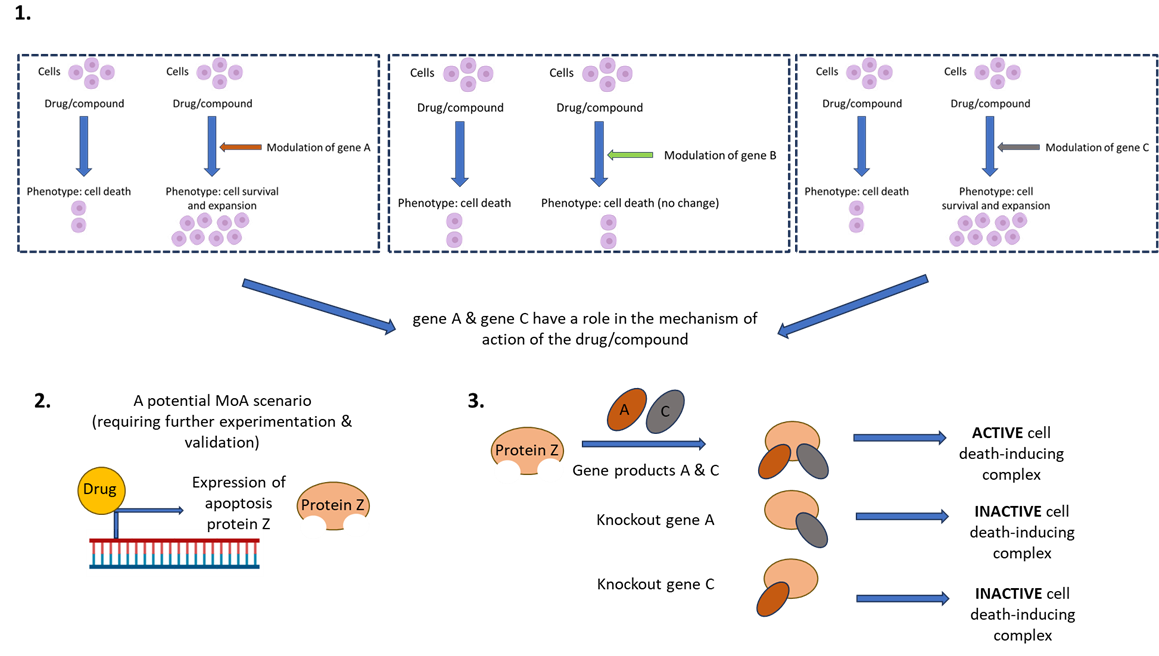
Figure 2. CRISPRko screen to investigate a hypothetical novel therapeutic compound. The compound induces cell death in a specific cancer cell type. If we want to discern the MoA of patient resistance to the activity of this compound, we can conduct a whole genome CRISPRko screen which will identify gene deletions which result in cell survival (rather than the intended cell killing by the compound). Further validation and investigation of these candidates might unveil the mechanism of drug function and indeed, drug resistance. (1) The varying phenotypes following differential genetic perturbation (in this example- gene knockout) and drug exposure. (2) Potential mechanism of the drug. (3) Mechanism of action of gene network and drug on phenotype.

